Intro to Bioinformatics Engineering, Part 1: The Purpose of Pipelines
When, why, and how to build a bioinformatics pipeline
This is the first article in our Intro to Bioinformatics Engineering series. In these articles, we will discuss some of the foundational blocks for building in bioinformatics. While these articles assume familiarity with programming and biology, they are written to be accessible for folks new to this field. We hope you enjoy!
Introduction: Hiking or Driving
Analyzing new data is a bit like hiking in unmapped woods. You’re hoping to find something, but you might not know what that something is or where it will be. Armed with a compass and a blank map (or whining laptop and Jupyter Notebook), you set out to explore. If you find something interesting you might make a note, and if you find something exciting you might start a new trail.
Running a production bioinformatics pipeline is more like driving on a road. You (hopefully) know where you are going, and it will be faster than walking. Most drivers on the road are not the engineers who built it. Your priorities differ from a hiker’s; it’s more important to travel quickly and reliably than to investigate something off the route.
For a traveler, the tradeoffs between trail and road are usually clear. But the considerations become far more complicated if, instead, you are a civil engineer.
In this post, I will discuss what makes an amazing bioinformatics engineer, what they might consider when “city planning,” and a few of the most common patterns we have seen for developing a successful bioinformatics system.
Why and When to Build a Pipeline
“Analysis” and “pipeline” can mean many things and are sometimes used interchangeably. Without claiming that these are the only definitions, for the sake of this article, they mean the following:
Analysis (hiking): Code that is written and run in a single setting. Examples: analyzing data in a Jupyter Notebook, editing and running an R script locally.
Pipeline (driving): Code that is written ahead of time and used at runtime. Examples: running a Python script with Snakemake, using 10x’s Cell Ranger Count, running Illumina’s BCL2FASTQ.
End-to-end data processing often requires a combination of analyses and pipelines.
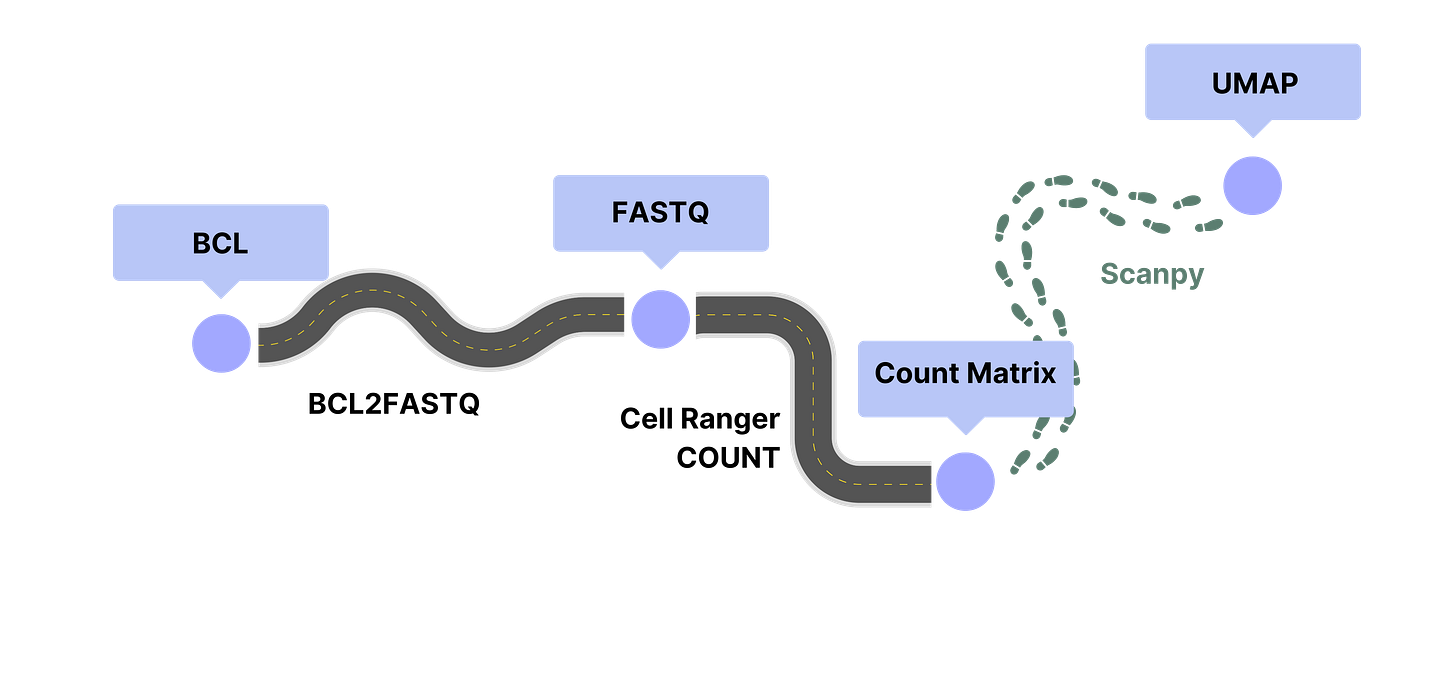
As an example, processing single cell transcriptomics data often begins with the raw output of a DNA sequencer (BCL).
The BCL2FASTQ pipeline converts the raw observations of the DNA sequencer into human-readable files containing DNA sequences and quality scores (FASTQ).
The Cell Ranger COUNT pipeline groups the reads in the FASTQ file by cell and by gene to create a table of gene counts per cell (Count Matrix).
To understand and visualize this data, someone does an analysis of the count matrix using the Scanpy package in a Jupyter Notebook. After gaining understanding, they create a visualization (UMAP) of the data to communicate their insights to others.
This example captures a few benefits of pipelines:
Shareable. A well-built pipeline can be written by a domain expert and then run by anyone. Illumina and 10x developed BCL2FASTQ and Cell Ranger COUNT respectively, enabling many scientists to analyze data without developing them from scratch.
Consistent. If two different people analyze the same dataset, they will likely make slightly different choices. This may be as superficial as different line colors in a graph or as significant as choosing different statistical tests. Using versioned pipelines makes processing consistent and results comparable, and may facilitate interoperability with other systems.
Testable. Almost everyone who works with sequencing data has used the output of BCL2FASTQ and trusts that the pipeline worked correctly. Because the pipeline has been benchmarked and tested by many use cases, it is now highly reliable and trustworthy.
Scalable. Suppose someone asks you to process thousands of BCL files with the process above. Though the analysis step will likely stay time-consuming, the pipeline steps can be run in parallel and automated.
Reproducible. If you capture the input, environment, and pipeline version you can reproduce a pipeline run. Analyses may also be reproducible, but it can require meticulous manual documentation.
But there are also advantages to analyses:
Flexible. Flexibility is naturally a tradeoff with consistency. Everyone loves being able to change something quickly. No one likes when a coworker’s quick change introduces a bug and blocks operations. If you want to move fast (and break things), an analysis is the answer. In an analysis, it is much easier to pull in additional files and packages on the go.
No “Upfront Cost”. A road must be planned and paved before others can drive on it. Similarly, building a pipeline requires more up-front work than using an analysis. An analysis only needs to support one exact use case; a pipeline should be designed to handle a range of values and inputs.
No Maintenance Cost. Roads must be maintained to be usable. Pipelines are not built in a vacuum; requirements, inputs, and infrastructure will change, and the pipeline will need to change to keep up. If you live somewhere with cold winters, imagine driving on a road that has not been repaved after a few freeze-thaw cycles. It is similarly unpleasant to use a pipeline that has not been updated after a faster or better tool became available.
In the example above, when would you replace the Count Matrix analysis with a pipeline? If you build a pipeline you only run once, you’ve wasted effort and slowed progress. If you use analyses hundreds of times, the results may be too inconsistent to analyze in aggregate.
How to weigh the tradeoffs depends on your priorities and capabilities. The faster you can write a pipeline and the more easily you can maintain it, the sooner you should do so. If you plan to change requirements frequently, it may be better to use analyses for longer. By combining both pipelines and analyses in your workflows, you can have the best of both worlds.
Common Pipeline Patterns
If you’re working with a complex analysis, it may be unclear if and how you should convert it to a pipeline. Here are a few of the most common patterns we have seen for creating pipelines.
The Last Mile
Many people experience the last-mile problem while commuting: the train almost takes you to the office but doesn’t quite get you there, or you park in a structure a few blocks from your workplace.
Here are some symptoms that your workflow is suffering from the last mile problem:
You feel like it’s “almost” a pipeline, but you want to be able to edit the code for the last step regularly.
There is an “intermediate file” that your pipeline produces.
You find yourself opening that intermediate file in a Notebook for an additional analysis.
Your script may take that intermediate file as an optional input, and when provided, a significant section of code is bypassed.
You are frequently asked to reprocess data with a small change to a final step.
What is often happening in this case is that the analysis begins with data cleaning/preprocessing, but ends with visualization/postprocessing. While the first step is fairly stable, the second step undergoes frequent changes and development.
Solution:
Create a pipeline for the “preprocessing” step.
If there is an intermediate file generated, use this as the checkpoint. If your pipeline has clear preprocessing logic but no intermediate file, think about a way you can create a standardized file to write out after the preprocessing. If you’re using Python, an easy option may be to pickle the intermediate object.
The “postprocessing” step may either be a pipeline or an analysis.
This depends on how frequently you are updating this section.
A common pattern is to start with an analysis and then convert it to a pipeline when it is stable. If you need to do a bespoke analysis, you can always open your intermediate file in a Jupyter Notebook. More on that below.
Multiple Destinations
“Multiple Destinations” is very similar to “The Last Mile.” The key difference is whether the pipeline always produces outputs of the same general type. Here are a few other symptoms of the Multiple Destinations pattern:
There are large sections of code that only run when certain conditions are met. For example, perhaps there is a graph that is only produced for data from a specific CRO, or there are different QC checks for different organisms.
You are frequently changing the script for certain types of input, but for other types of input you can run the analysis as-is.
Solution:
Go through the code, line by line if necessary, and determine which sections run for ALL inputs.
Separate this logic into a separate function or pipeline. This may take some refactoring. This will also require determining or creating an intermediate file, similar to the Last Mile pattern.
Examine the code that does not run for all inputs. What determines when each of these runs? Is it the data type of the input? Is it a property, like the organism? Is it whether the raw data passes or fails a QC check? Make a list of these categories.
Look at the processing and output for each category. You may find that some categories have a very established analysis that can be converted to a pipeline while others are still exploratory.
Example:
Suppose you are processing videos of zebrafish to study behavior. You had been doing experiments to study feeding behavior, and now you are beginning to study social behavior. Your script always begins by converting the video into wireframe models of the fish. Then, depending on the type of experiment, either feeding or social behavior metrics are extracted from the wireframes.
Solution:
Create a pipeline that takes any video and produces wireframes. Create a second pipeline that takes wireframes and calculates feeding metrics. Since you are still developing the social behavior experiments, continue to analyze the data in a Jupyter Notebook until the protocol is refined. Now, if your coworkers want to start analyzing a new type of behavior they can use your preprocessing pipeline!
The First Mile
The most common story for The First Mile pattern is the following:
You’d like to do an analysis involving many datasets. These datasets are all of the same general type, but they come from multiple collaborators, CROs, instrument types, public databases, or protocols. You are frequently editing your script because it does not quite work for a new dataset, or you are manually tweaking the dataset to make it a valid input to your pipeline.
The solution is basically the same as for the Multiple Destinations Pattern, but reversed. First, determine the part of your script that runs for all data, and separate it into a pipeline. The input to this pipeline may be obvious, or it may take a little creativity.
Once you have decided on the input to the pipeline, your next goal becomes clear. For each new raw dataset, convert the dataset into the input. Depending on your data, this might mean writing an analysis for every dataset! Or maybe you will find that some datasets are similar enough to create a preprocessing pipeline. Either way, you do not need to edit the processing pipeline for each new dataset.
Pipeline Building is One Part of Bioinformatics Engineering
When should a city build a new highway? Where should the road go? Is a highway worth the investment? Will the road be able to widen and scale when the city grows in the future?
Similar to civil engineering, great bioinformatics engineering means building reliable, scalable, and ready-for-change systems.
Bioinformatics engineers are not the only ones who do bioinformatics engineering. Bioinformatics engineering is often a critical skill for computational biologists, data engineers, software engineers, scientists, and research engineers.
Bioinformatics engineering overlaps with computational biology and software engineering. While it’s common for one person to wear multiple hats, it's helpful to consider these as separate skills.
Advice for Teams
Great bioinformatics engineers are often people who love working in a collaborative environment. Commonly, the creator of the data, developer of the algorithm, and stakeholder for the results are three different people. Here are a few thoughts on effective collaboration with these groups:
If you ask someone where they would like a road, they will likely ask for it to be a straight line from their starting point to their destination. Unfortunately, if this was done for everyone, the countless overlapping roads would be nearly impossible to maintain or change. Requests from users are often focused on the fastest solution to the current problem; bioinformatics engineers need to balance this with efficiency and sustainability. It’s important that a team is structured to allow scientists and engineers to collaborate on project requirements.
When developing a new pipeline, it’s important to have clearly communicated requirements from the teams that will use it. I know not every engineer loves documentation, but having a shared design doc can prevent timelines from slipping due to misunderstood requirements. Consider versioning the document, and having 1-2 stakeholders “sign off” on every new version.
Be careful not to convert an analysis into a pipeline too early; you wouldn’t want to build a road that never gets used. A good rule of thumb is to convert an analysis to a pipeline only after it has been run at least five times without significant changes. This has the additional benefit of providing testing data to validate the pipeline.
Bioinformatics is a BLAST
I had a great time writing this, and I hope you enjoyed reading it! We have more posts planned for our Intro to Bioinformatics Engineering Series including guides for leveraging essential tools, tips for being ready-to-scale, example developer workflows, and more. Please subscribe below if you would like to be notified, or visit us here for more bioinformatics tips.
Emily Damato is the CEO and Co-Founder of Mantle. Her favorite organism is Drosophila melanogaster.







Thank you for such clear explanation! The diagrams and the analogies were quite helpful.